ArrayCluster provides the user a number of available options for data preprocessing
including gene filtering and data normalization. In sequel, the details
will be described.
Gene Filtering
Although microarray dataset contains a large number of genes, a part of
genes are typically excluded during the expression profiling. This process,
i.e. Gene Filtering, is aimed at removing the undesirable-genes that contain outliers and
too much missing expression values, and that do not exhibit variability
across tissue samples. To this end, the ArrayCluster offers a number of
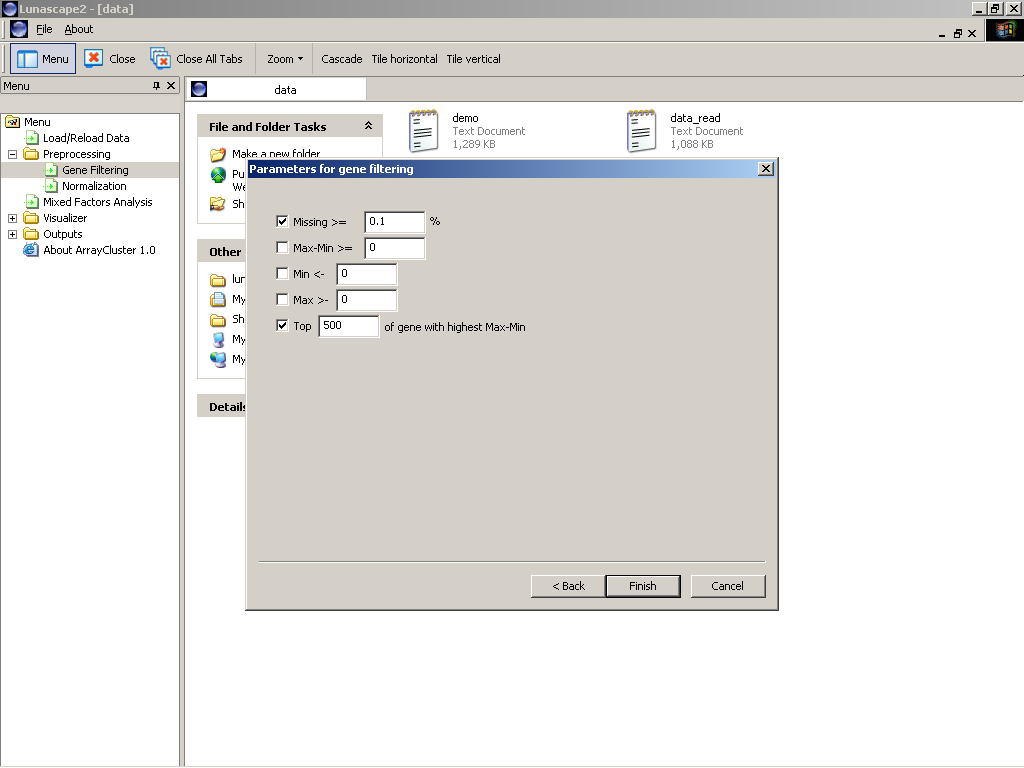
options. Tracing from "Preprocessing" to "Gene Filtering",
the following questions will be asked (as shown in Figure 1):
- Missing >= [X] %
Checking this dialog removes genes that contain missing values more than X %. - Max-Min >= [X]
This removes all genes that the differences between maximum and minimum expression values are less than X. - Min <= [X] / Max >= [Y]
This remove all genes that contain outliers below (or above) a thresholds X (or Y) a user specifies. - Top [X] of genes with the highest Max-Min
This option select genes to be top X of the highest maximum minus minimum values.
- --- SELECTED OPTIONS IN GENE FILTERING ---
* MISSING VALUES WITH MORE 1.0 %
* TOP 500 GENES WITH HIGHEST MAX-MIN
- * NUMBER OF SELECTED GENES 500
-----------------------------------------------------------
FILTERED DATA --->
C:\ArrayCluster\extents\lunamacplugin\data\data_read.txt
-----------------------------------------------------------
Fortran Pause - Enter command<CR> or <CR> to continue.
If not so, any error messages might be emerged in this window.
List of the selected gene filtering options can be checked through the process viewer. In this example, missing cut with 1% and the largest deviance 500 genes selection were chosen. The number of selected genes, in this case, 500, is also noted. If users would like to reduce or increase the number of genes involved in the following profiles, turn to reload data file step and again proceed to the gene filtering process.
The filtered data file will be created at \C:ArrayCluster\extents\LunamacPlugin\data\ data_read.txt. By the inspection, user can also check whether the filtering process has been accurately completed. To close the DOS window, push Enter key.
Figure 1: Gene filtering wizard
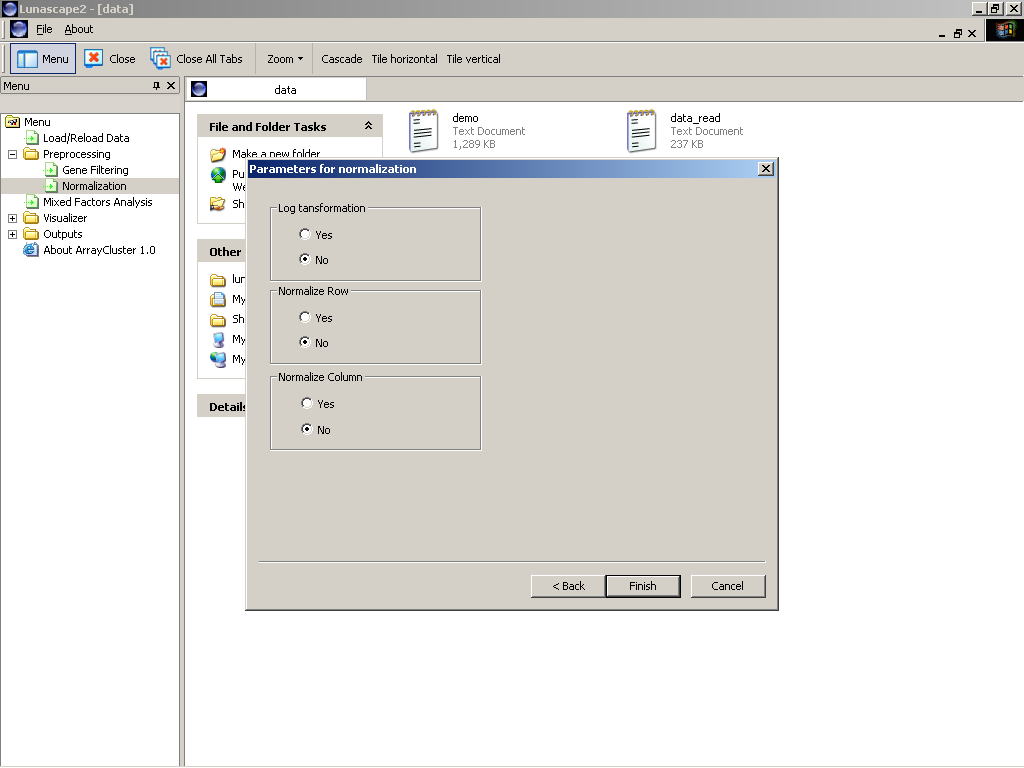
Normalization
DNA microarray experiments often have systematic bias, for instance, heterogeneity in the scaling and origin, due to the experimental design. These biases should be possibly adjusted before proceeding to the downstream analysis. In microarray studies, this step is referred to as the Normalization.
ArrayCluster 1.0 offers some available normalization methods. Proceeding to "Normalization" menu, then user can see the normalization wizard (Figure 2) in which the following options are listed:
- Log Transformation:
Taking a natural logarithm of loaded values. All values to be transformed by Log (x) must be positive numeric numbers. - Normalize Array/Gene:
Shifting the origin of gene expression values and rescaling them in each row (column) to have mean zero and unit variance.
After checking any dialogs and clicking "Finish", normalization.exe will run on. If the process is completed, the following messages will appear in the DOS-prompt window:
- --- SELECTED OPTIONS IN NORMALIZATION ---
* NORMALIZE ARRAYS
* NORMALIZE GENES
-----------------------------------------------------------
NORMALIZED DATA --->
C:\ArrayCluster\extents\lunamacplugin\data\data_read.txt
-----------------------------------------------------------
Fortran Pause - Enter command<CR> or <CR> to continue.
If not so, some errors are alerted by this window. For example, augment of the logarithmic function should be a positive real value. If user selected Log Transformation option in spite of that the loaded data contains any negative gene expression values, the following error messages will appear in the DOS-window:
- --- SELECTED OPTIONS IN NORMALIZATION ---
* LOGARITHMIC TRANSFORMATION
-----------------------------------------------------
WARNING: DOMAIN ERROR IN LOGARITHMIC TRANSFORMATION
***** RESELECT OPTIONS ! *****
-----------------------------------------------------
-----------------------------------------------------------
NORMALIZED DATA --->
C:\ArrayCluster\extents\lunamacplugin\data\data_read.txt
-----------------------------------------------------------
Fortran Pause - Enter command<CR> or <CR> to continue.
Figure 2: Normalization wizard