Clicking "Mixed Factors Analysis" in menu, run the input parameter wizard. User is then required to specify the following parameters:
- Range of factor dimensions (number of module transcriptional) taken into
considerations:
For example, if user sets the factor dimensions ranging from Max=8 to Min=5, the ArrayCluster compute the corresponding BIC scores, and then select an optimal factor dimension, say Factor Dimension = 7, that attains the minimum BIC. The following results, e.g. clustering, module identification and so on, corresponds to those given by an optimal model, i.e. Factor Dimension=7. If user takes into consideration just uni-model, e.g. Factor Dimension=4, input "Factor Dimensions" equal to Max=4 and Min=4. Note that user must not enter any inconsistent values such as Max < Min, Max=0 and Min=0. The current version 1.0 places a 20 upper-limit on the maximum factor dimension.
- Range of the number of clusters taken into considerations:
As in the case of the determination of factor dimension, the ArrayCluster searches an optimal number of clusters based on the BIC scores. For a specified candidate ranging Min <=#.Cluster <=Max and Min <=Factor Dimension<=Max, the program finds a most suitable combination, then return some outputs corresponding to the selected combination. Operation procedures and notandums are same with those of Factor Dimension.
- Initialization options:
For the mixed factors model, the likelihood equations to be solved have multiple roots. Hence, the computation of EM algorithm should be repeated several times with a wide range of the initial parameters. The current version implements the randomized initialization for the determination of starting paramters with several times user specifies.
- Common Covariance Matrix Options:
Mixed factors model assumes that the q-dimensional factor vector is distributed according to the mixture of the Gaussian distributions having the diagonal covariance matrices, V_g=diag(v_1g,····, v_qg), for g=1,····,G. ArrayCluster 1.0 equips the option for the use of isotopic covariance matrices, i.e. V_g=vI for all g.
- Number of Relevant Genes:
As was already mentioned, the mixed factors analysis identifies total 2×(Factor Dimension) gene modules that are relevant to group structure of the gene expression signatures. In ArrayCluster, user can select the number of genes selected as members in one module through this option dialog.
Implementation
After specifying all parameters and pressing the "Finish" button, the Mixed_Factors_Analysis.exe runs on. If the process is completed, the messages shown in below appear on the DOS-windows.
At first, experimental parameters specified by user are notified, for example, Max and Min factor dimension, number of relevant genes to be selected.
Going a layer than that, user can view the process of EM algorithm for a combination of each factor dimension and number of clusters and initial parameters. The statements such as "* -C" or "* -OF" represent states of the convergence for the EM algorithm, "in convergence" or "occurrence of over-fitting", respectively.
Furthermore, a most suitable combination of factor dimension and the number of clusters are also shown, in this example, the optimal factor dimension and the number of clusters are equal to 7 and 5.
Subsequently, the calibrated clusters are shown. Each cluster is separated with the blank lines. Note that all samples in one clusters are ordered according to the degree of the belongings that are measured by the magnitude of Maharanobis distance between the sample points and the corresponding group centeroid.
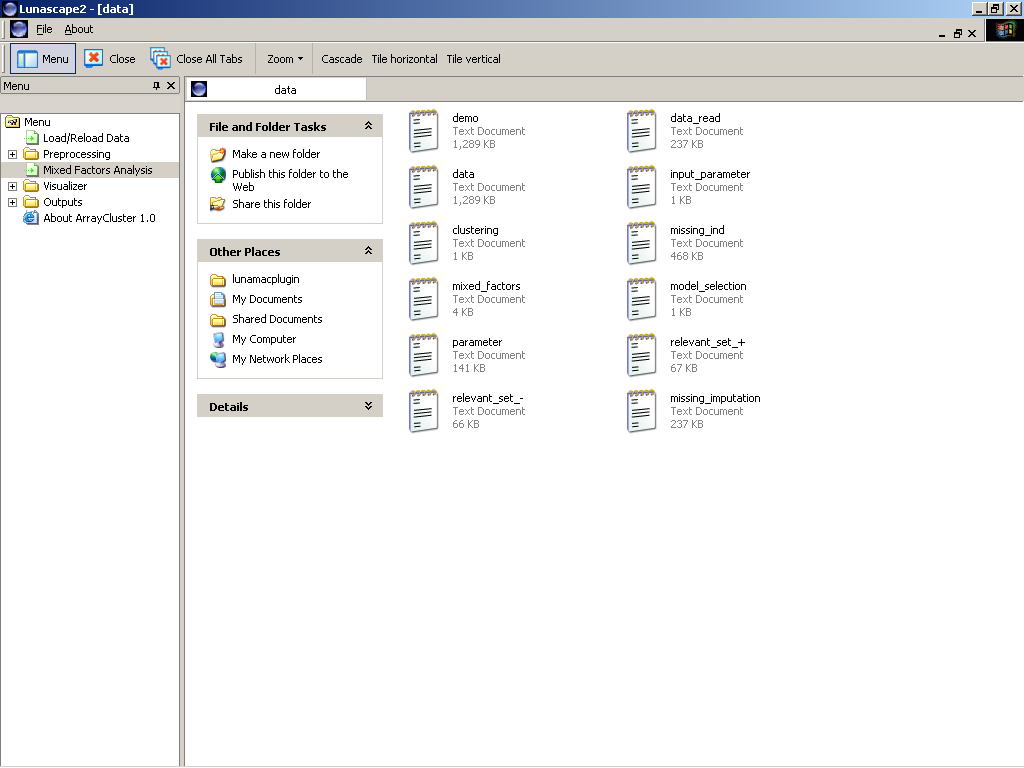
Finally, the created six files that summarize some results of the mixed factors analysis and the folder locations are notified.. Description of the six files are in turn given through this tutorial, e.g. Model Selection, Clustering, Data Compression and Relevant Module Detection.
Figure 1: Output files creadted by the mixed factros analysis
Messages appeared in DOS window (Mixed_Factors_Analysis.exe)
- --- SELECTED OPTIONS FOR MIXED FACTORS ANALYSIS ---
* Max_Factor 8
* Min_Factor 6
* Max_Num_Clusters 6
* Min_Num_Clusters 4
* Num_Iterations_EM 3
* Common_Covariance N
* Num_Relevant_Feature 20
-----------------------------
PROCESS VIEW OF EM ALGORITHM
FACTOR DIM. 6 NUM.CLUSTERS 4
* -C
* -OF
* -C
FACTOR DIM. 7 NUM.CLUSTERS 4
* -OF
* -OF
* -OF
* -C
* -C
* -C
FACTOR DIM. 8 NUM.CLUSTERS 4
* -C
* -C
* -C
FACTOR DIM. 6 NUM.CLUSTERS 5
* -C
* -C
* -C
FACTOR DIM. 7 NUM.CLUSTERS 5
* -C
* -OF
* -OF
FACTOR DIM. 8 NUM.CLUSTERS 5
* -C
* -OF
* -C
FACTOR DIM. 6 NUM.CLUSTERS 6
* -OF
* -OF
* -C
FACTOR DIM. 7 NUM.CLUSTERS 6
* -OF
* -OF
* -OF
* -C
* -OF
* -OF
FACTOR DIM. 8 NUM.CLUSTERS 6
* -OF
* -C
* -OF
-----------------------------
--- SELECTED MODEL ---
FACTOR DIM. 6 NUM.CLUSTERS 4
MIN. BIC -173757.9536
--- CLUSTERING ---
ex2 2.5143
ex56 3.0997
ex53 3.3880
ex17 3.6238
ex54 3.6811
ex61 3.7153
ex32 4.0781
ex21 4.3215
ex15 5.0945
ex16 5.4574
ex51 5.5414
ex55 6.4377
ex4 6.4684
ex1 7.3439
ex34 2.6977
ex26 2.8049
ex27 3.3646
ex28 3.4936
ex30 4.3155
ex40 4.5669
ex31 5.3043
ex25 5.9297
ex33 6.1687
ex29 8.3128
ex24 10.7974
ex58 2.7702
ex6 2.8320
ex60 2.9380
ex5 3.4753
ex45 3.7771
ex11 3.8016
ex13 4.0272
ex8 4.3992
ex12 4.5219
ex9 4.6122
ex57 4.7551
ex7 4.7553
ex59 5.2079
ex10 5.8650
ex3 6.6773
ex44 8.2157
ex62 8.8822
ex63 16.1418
ex52 1.0337
ex20 3.1033
ex50 3.5920
ex22 3.6395
ex43 3.8607
ex37 3.8875
ex39 3.9114
ex36 4.1459
ex48 4.2865
ex41 5.2053
ex23 5.6382
ex47 5.7172
ex46 6.0053
ex18 6.9978
ex19 7.0149
ex49 7.0154
ex35 7.3917
ex38 8.6547
ex42 8.6757
ex14 9.2254
OUTPUT FILE DESCRIPTION
-----------------------------------------------------------
* BIC Scores --->
C:\ArrayCluster\extents\lunamacplugin\data\model_selection.txt
* Clustering --->
C:\ArrayCluster\extents\lunamacplugin\data\clustering.txt
* Factor Scores --->
C:\ArrayCluster\extents\lunamacplugin\data\mixed_factors.txt
* Relevant Positive Gene Sets --->
C:\ArrayCluster\extents\lunamacplugin\data\relvant_set_+.txt
* Relevant Negative Gene Sets --->
C:\ArrayCluster\extents\lunamacplugin\data\relvant_set_-txt
* Estimated Parmeters --->
C:\ArrayCluster\extents\lunamacplugin\data\parameters.txt
* Missing Imputation --->
C:\ArrayCluster\extents\lunamacplugin\data\.missing_imputation.txt
-----------------------------------------------------------
Fortran Pause - Enter command<CR> or <CR> to continue.